Will kinase domain mutations dictate the terms…
CC BY-NC-ND 4.0 · Indian J Med Paediatr Oncol 2013; 34(03): 151-153
DOI: DOI: 10.4103/0971-5851.123707

Publication History
Article published online:
19 July 2021
© 2013. Indian Society of Medical and Paediatric Oncology. This is an open access article published by Thieme under the terms of the Creative Commons Attribution-NonDerivative-NonCommercial-License, permitting copying and reproduction so long as the original work is given appropriate credit. Contents may not be used for commercial purposes, or adapted, remixed, transformed or built upon. (https://creativecommons.org/licenses/by-nc-nd/4.0/.)
Thieme Medical and Scientific Publishers Pvt. Ltd.
A-12, 2nd Floor, Sector 2, Noida-201301 UP, India
Optimization of chronic myeloid leukemia-chronic phase (CML-CP) treatment is a continuous process. Although imatinib is a major breakthrough (that too, almost a decade ago), every third or fourth patient has to come off this therapy due to resistance or intolerance.
BCR-ABL kinase domain (KD) mutations account for 50-90% of the imatinib resistance observed in patients of CML-CP.[1] Over 90 mutations have been identified so far [Figure 1]. These mutations have variable frequency and confer variable degree of resistance to imatinib. Experience with second generation tyrosine kinase inhibitors (2G-TKI) i.e., dasatinib and nilotinib has shown much narrower spectra of mutations and fortunately these spectra are non-overlapping, the T315I (the gatekeeper mutation) being the unique exception.
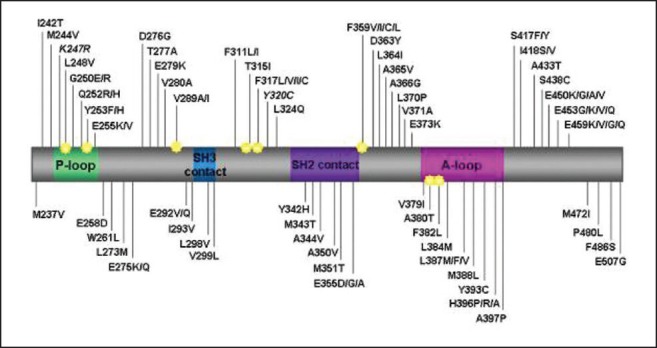
| Fig. 1 The BCR-ABL kinase domain mutations in patients with imatinib resistance. (P-loop, phosphate binding loop; A-loop, activation loop [reproduced with permission from Editor, blood, pre-published online May 19, 2011])
It is important for clinicians to know the timing for performing mutation analysis while the laboratory person has to know how to perform it. Lastly, hematologists and medical oncologists have to translate the results into clinical practice.
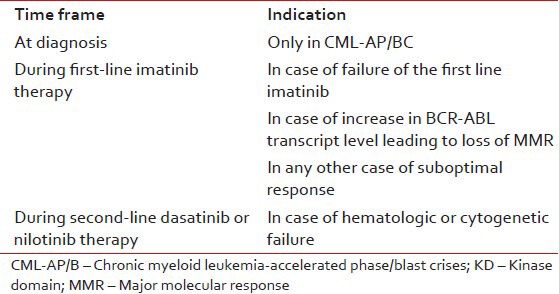
In CML-CP, patients receiving imatinib first-line, mutation analysis is recommended only in case of failure or suboptimal response using European LeukemiaNet (ELN) criteria. In imatinib-resistant patients receiving an alternative TKI, mutational analysis is recommended in case of hematologic or cytogenetic failure, once again as defined by ELN. Mutation analysis must not be performed at diagnosis, unless patient has CML-accelerated phase/blast crises [Table 1 and Figure 2].
Table 1
Recommended indications for performing BCR-KD mutation analysis
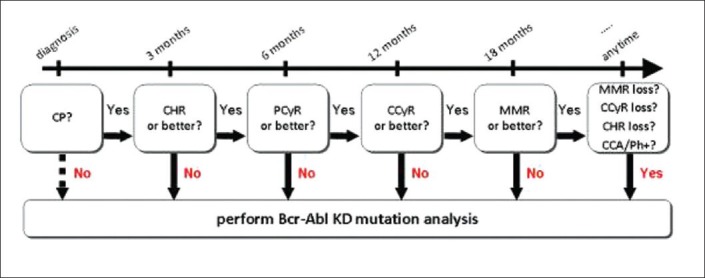
| Fig. 2 Flow-chart summarizing when mutation analysis is recommended in chronic myeloid leukaemia-chronic phase patients treated with imatinib first-line. (CP – Chronic phase; CHR – Complete hematologic response; PCyR – Partial cytogenetic response; CCyR – Complete cytogenetic response; MMR – Major molecular response; CCA/Ph + – Clonal chromosomal abnormalities in the Ph + clone
Mutation analysis is now being performed in a growing number of laboratories. However, there is still considerable confusion as to the techniques to be used and how the results should be interpreted. The recommended methodology is direct sequencing, although, it may be preceded by screening with other techniques like denaturing-high performance liquid chromatography (D-HPLC). D-HPLC is a straightforward and high-throughput tool to pre-screen for sequence variations, resulting in a great reduction of the number of samples that need to be sequenced. Other methods having higher sensitivity like fluorescent allele-specific polymerase chain reaction detect rarer mutations; however, the results do not correlate with the clinical picture of imatinib resistance. It is possible that these cells with rare mutations are not capable of sustaining long-term hematopoiesis and are effectively outnumbered by the unmutated ones.
In this issue, Srivastava and Dutt have presented an overview of their experience regarding detection of BCR-ABL KD mutations using direct sequencing in Indian CML patients on imatinib therapy.[2] According to published literature, although, over 90 mutations have been described, only 9 of these are common and account for almost 85% of mutations.[3] Similarly, Srivastava and Dutt in the present publication found nine mutations accounting for over 85% of total mutations seen in their laboratory. They have also compared distribution and relative frequency of mutations found in their lab with those reported by GIMEMA working party on CML. Significant differences were observed with higher frequency of mutations noted in Indian series at aminoacids T315, F359 and G250.
In all the cases outlined above, a positive result is an indication for therapeutic change. The options include-increasing imatinib dose,[4] switching to 2G TKI dasatinib or nilotinib,[5,6] moving to allogeneic stem cell transplantation[7] or enrolling patient in ongoing clinical trials having investigational compounds. In this issue, Rajjapa et al. have published their experience with KD mutations and responses to imatinib dose escalation in patients of CML-CP resistant to standard dose of imatinib.[8] Authors accept that due to financial constrain, escalated dose imatinib is the most practical and often, the only option for the majority of patients in India. In their series, unfortunately, the gatekeeper mutation, T315I accounted for almost 1/3rd of all the mutations and that is bad news as at the moment, except transplantation, no treatment is available to such patients. In this study, as a higher percentage of patients had hematological failure (rather than cytogenetic failure), the response to imatinib dose escalation was inferior to that reported in the literature. This further goes to show the importance of regular cytogenetic and molecular monitoring during follow-up of patients on imatinib therapy.
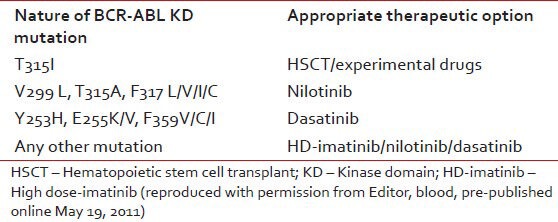
Certain specific mutations weigh on TKI selection [Table 2]. In case of T315I mutation, which is highly resistant to imatinib, dasatinib and nilotinib, there are no pharmacologic opportunities other than investigational compounds or going for allogeneic stem cell transplantation. Different TKIs exhibit different IC50 values and it is tempting to let the TKI choice result from the IC50 value for a particular mutation, i.e., selecting the TKI with the lower IC50. However, clinicians must remember that IC50 is an imperfect tool to guide optimal selection of 2G TKI. It does not account for a spectrum of factors determining the effective intracellular drug concentrations such us absorption, metabolism and distribution to target compartments, transport and excretion.
Table 2
Most appropriate alternative therapeutic options based on the BCR-ABL KD utation status
Lastly, it must never be forgotten that besides BCR-ABL KD mutations, there are other mechanisms of imatinib resistance. In a proportion of cases, more than one factor may exist. In fact, sometimes, mutations may be simple “bystanders.” Still, the presence of mutation can never be overlooked as mutation suggests genetic instability and genetic instability is the engine of disease evolution toward a more aggressive phenotype.
Footnotes
Source of Support: Nil.
Conflict of Interest: None declared.
REFERENCES

| Fig. 1 The BCR-ABL kinase domain mutations in patients with imatinib resistance. (P-loop, phosphate binding loop; A-loop, activation loop [reproduced with permission from Editor, blood, pre-published online May 19, 2011])

| Fig. 2 Flow-chart summarizing when mutation analysis is recommended in chronic myeloid leukaemia-chronic phase patients treated with imatinib first-line. (CP – Chronic phase; CHR – Complete hematologic response; PCyR – Partial cytogenetic response; CCyR – Complete cytogenetic response; MMR – Major molecular response; CCA/Ph + – Clonal chromosomal abnormalities in the Ph + clone


 PDF
PDF  Views
Views  Share
Share

