Personalized medicine: FAQs
CC BY-NC-ND 4.0 · Indian J Med Paediatr Oncol 2010; 31(02): 72-74
DOI: DOI: 10.4103/0971-5851.71661

|
Publication History
Article published online:
19 November 2021
© 2010. Indian Society of Medical and Paediatric Oncology. This is an open access article published by Thieme under the terms of the Creative Commons Attribution-NonDerivative-NonCommercial-License, permitting copying and reproduction so long as the original work is given appropriate credit. Contents may not be used for commercial purposes, or adapted, remixed, transformed or built upon. (https://creativecommons.org/licenses/by-nc-nd/4.0/.)
Thieme Medical and Scientific Publishers Pvt. Ltd.
A-12, 2nd Floor, Sector 2, Noida-201301 UP, India
1. WHAT IS PERSONALIZED MEDICINE?
Currently, an individual is treated for a particular disease, based on the data obtained from thousands of other patients. A physician presumes that the new patient would also respond to the drugs which he prescribes. However, not always, do these presumptions turn out to be correct. Additionally, there could be instances wherein the treatment may produce severe side-effects, sometimes even leading to fatal consequences. Again, while dose-related side-effects are easier to predict, others may not be so.
The genomic revolution now promises to help identify the most appropriate drug for a disease for a given individual, not only ensuring activity but also avoiding those that can cause toxicity. Thus, the medical care is now tailored to an individual’s characteristics (genomic information) and can be called as Personalized Medicine.
2. WHAT IS THIS GENOMIC INFORMATION?
Genome can be defined as the sum of all the genetic information in a given individual. We all know that the alphabets for the DNA are Adenine, Guanine, Cytosine and Thymine (A, G, C, T). It is the way these alphabets are stringed together which determines the gene sequence. When a particular gene needs to be expressed as a protein, its DNA sequence is read out as a messenger RNA (mRNA), which in turn moves from the nucleus of the cell to the cytoplasm wherein it is then translated into protein [Figure 1]. In the mRNA, Uracil replaces Thymine. The machine which does the translation of mRNA to protein is called ribosome and it does this by reading three alphabets at a time (called as triplet codon). Each codon codes for an amino acid which are the building blocks for the proteins – thus AUG codes for the amino acid Methionine, ACG codes for Threonine, AUC codes for Isoleucine, AAU for Asparagine and so forth. There are also codes for starting the translation (initiation codon, which is AUG coding for Methionine) and stopping the translation (stop codons which are UGA, UAG, UAA) of the mRNA, which ensures that the right length protein is synthesized. Currently, it is presumed that there are about 25,000 genes in the humans. However, a large number of genes have different/alternate start codons which can result in different sized protein variants (transcript variants), giving rise to additional complexity.
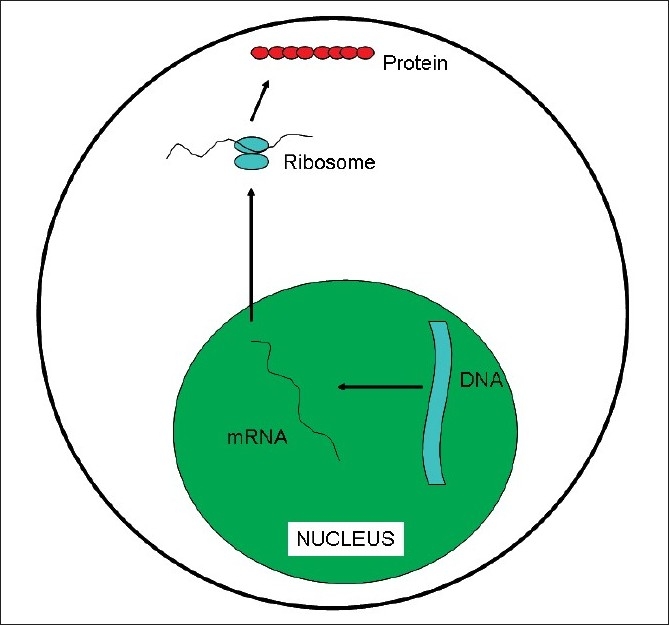
| Figure 1:DNA to RNA to protein
As humans, all of us have a similar genetic code, which is nearly 99.9% similar. The 0.1% difference is what makes us different, you being you and your friend being him. These differences are due to change in the DNA sequence, sometimes a mere change of one alphabet. This is called as Single Nucleotide Polymorphisms or SNPs. The effect of one alphabet change can be variable; some could have a profound change in the meaning/function while others may have minimal or no effect, as shown below:
e.g. Cattle – Battle; Tumour – Tumor
| DNA code | Amino acid |
|---|---|
| TGG | Tryphtopan |
| TCG | Serine |
| TTG | Leucine |
| TAG | STOP |
In the example shown above, a single alphabet change in the second position of the codon leads to changes in the amino acid and worse can lead to prematurely stopping the translation of the protein which is likely to be functionally inactive. The latter is called as Nonsense mutation while the others are called as missense mutations.
3. WHAT ARE THE EFFECTS OF THE GENETIC MUTATIONS/POLYMORPHISMS?
There are three possible effects on the protein function:
- no effect on the protein function or
- decreased efficiency of protein function or
- increased efficiency of protein function.
Let us take a gene which codes for a protein which is involved in the metabolism of certain cancer causing chemicals. This protein could be involved in breaking down the harmful cancer causing chemicals to harmless compounds which can then be excreted by the body [Figure 2]. If there is an SNP in the gene and if it alters the function such that this protein functions only at 50%-of its inherent efficiency, then the cancer causing chemicals will be present in the body for a longer duration, leading to an increased risk for the development of cancer; on the contrary, if the protein functions at 150%-efficiency, then the cancer causing chemical is rapidly broken down, resulting in a lower risk for cancer (this could explain why some smokers do not develop cancers).
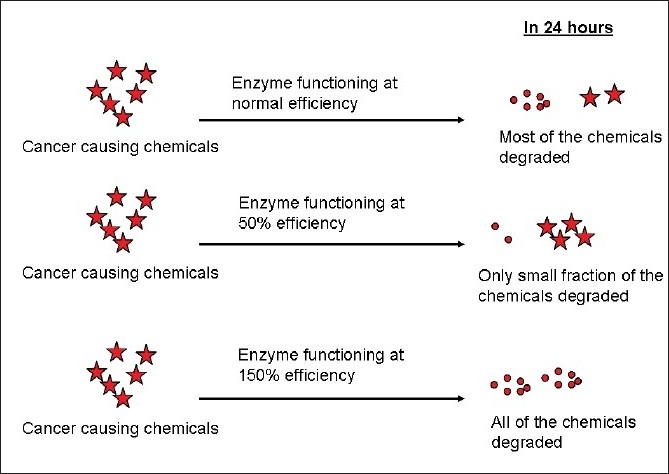
| Figure 2:Effect of SNP on protein function

| Figure 1:DNA to RNA to protein

| Figure 2:Effect of SNP on protein function


 PDF
PDF  Views
Views  Share
Share

