Next Generation Sequencing
CC BY-NC-ND 4.0 · Indian J Med Paediatr Oncol 2020; 41(03): 381-382
DOI: DOI: 10.4103/ijmpo.ijmpo_28_20
Next generation sequencing (NGS) has revolutionized the field of medicine and research and has improved on the principles of first-generation sequencing (Sanger Sequencing).
Publication History
Received: 27 January 2020
Accepted: 18 March 2020
Article published online:
28 June 2021
© 2020. Indian Society of Medical and Paediatric Oncology. This is an open access article published by Thieme under the terms of the Creative Commons Attribution-NonDerivative-NonCommercial-License, permitting copying and reproduction so long as the original work is given appropriate credit. Contents may not be used for commercial purposes, or adapted, remixed, transformed or built upon. (https://creativecommons.org/licenses/by-nc-nd/4.0/.)
Thieme Medical and Scientific Publishers Pvt. Ltd.
A-12, 2nd Floor, Sector 2, Noida-201301 UP, India
Next generation sequencing (NGS) has revolutionized the field of medicine and research and has improved on the principles of first-generation sequencing (Sanger Sequencing).
What Is Sanger's Sequencing?
It is the classical method of sequencing, i.e., technique used to determine the order of 4 bases. This involves the following steps [Figure 1].[1]
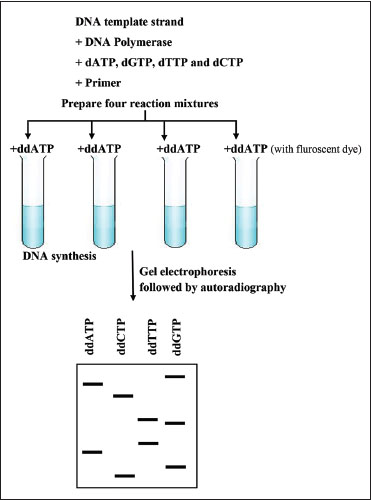
| Figure 1:Steps of Sanger’s sequencing
-
First, the DNA to be sequenced is amplified. These are then separated into single strands using heat. This leads to production of two strands (complementary strand and template strand). Using the 5' end of DNA in the template strand, a primer is annealed to it.
-
Primer DNA is dispersed into 4 vessels. DNA polymerase and all four dNTPs (dATP, dGTP, dTTP, dCTP) is added to all 4 vessels.
-
Next, 1 type each of the 4 ddNTPs is added to each vessel. These ddNTPs are specifically modified dNTPs which does not have a 3'hydroxyl group required for chain elongation. Hence, no chain elongation occurs when these enter the chain. Each ddNTP also has unique fluorescent dye attached to it, allowing for automatic detection.
-
Thus, varying length of DNA fragments is formed in each vessel which by gel electrophoresis followed by autoradiography its sequence is determined. Polyacrylamide gel is usually used for electrophoresis.
What Is Next Generation Sequencing?
Next generation sequencing is a newer high output DNA sequencing technology which utilizes massive parallel sequencing method. It sequences DNA similar to Sanger's sequencing but, it is a massively parallel method which allows millions of DNA fragments to be sequenced in a single run, in contrast to sanger sequencing which only produces one forward and reverse read only. Thus, in contrast to Sanger's sequencing, the speed and amount of DNA sequenced and data generated with NGS are greater with a much lower cost.[2] There are four sequencing approaches for NGS.[3] These are:
-
Pyrosequencing
-
Sequencing by synthesis: The most common method used now
-
Sequencing by ligation
-
Ion semiconductor sequencing
And based on sequence determination, NGS is divided into three types. These are:
-
Whole genome sequencing: Refers to sequencing of most of the DNA comprising both exon and intron.
-
Whole exome sequencing: Refers to sequence determination of exons (Exons are segments of DNA which encode proteins and are approximately 1%-of the whole genome).
-
Clinical exome sequencing: Refers to sequencing of morbid genes only.
Next Generation Sequencing in Oncology-What to Know?
-
Source of DNA: Can be blood (this is a sterile source of DNA, but an invasive procedure), buccal swab, saliva, hair follicle (this is a nonsterile source of DNA, with a potential risk of DNA of bacterial flora to be confused to host sequence). For patients undergoing allogenic hematopoietic transplant, blood sample contains donor's DNA. Hair follicle is the most reliable sample for recipient's genetic sequence. It can also be extracted from fixed samples (formalin fixed; paraffin embedded).
-
For research purposes, NGS can also be used for sequencing RNA and mitochondrial DNA
-
Which part of DNA to use…?
NGS can be used to sequence whole DNA, exome, or target gene panel. In clinical practice, we commonly use target gene panels for hereditary breast cancers and hereditary colon cancers. The results of these are reported as one of the following:
-
Pathogenic
-
Likely pathogenic
-
Benign
-
Unknown clinical significance.
There is a higher incidence of receiving a result of unknown clinical significance, as clinical implication of many variants is not known.
-
-
Clinical uses in oncology.
-
Cancer screening:
-
Breast cancer - BRCA1 and BRCA2, CHEK2, TP53 and ATM.
-
Colorectal cancer - APC, MLH1, MSH2, MSH6, MUTYH, TP53.
-
Familial acute leukemia syndrome - CEBPA, RUNX1, DDX41, MBD4, GATA2, TP53.
-
-
Cancer prognosis:
Acute myeloid leukemia-stratified into favorable risk, intermediate, and poor risk based on genetics.
-
Cancer treatment:
-
PARP inhibitors: Used when BRCA testing comes positive.
-
NGS can also be used to detect tumor mutation burden, which is useful in deciding the therapy in many cancers.
-
-
As a last ray of hope:
Gene panel tests: MSK-IMPACT and F1CDx-in recurrent and metastatic setting for solid tumors and metastases of unknown primary, helping in therapeutic decision.[4]
-
Limitations
This platform may not be useful for large chromosomal changes such as chromosomal copy number changes, deletion, insertions, and translocations. In these other diagnostics such as cytogenetics, fluorescencein situ hybridization, microarray may be useful. Cost and longer time are the other technical limitations
Future Implication
Advances in NGS have catalyzed progress in countless areas of cancer research, heralding a way toward personalized and precision medicine in its management. Thus, NGS may play a very important role in future in above and may even become a part of standard baseline investigation.
Acknowledgments
We are grateful to Dr. Sujith Kumar, consultant medical oncologist at Aster MIMS - Kannur for his continuous guidance throughout the study.
Conflict of Interest
There are no conflicts of interest.
References
- Sanger F, Coulson AR. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol 1975; 94: 441-8
- Grada A, Weinbrecht K. Next-generation sequencing: Methodology and application. J Invest Dermatol 2013; 133: e11
- Heather JM, Chain B. The sequence of sequencers: The history of sequencing DNA. Genomics 2016; 107: 1-8
- Kamps R, Brandão RD, Bosch BJ, Paulussen AD, Xanthoulea S, Blok MJ. et al. Next-generation sequencing in oncology: Genetic diagnosis, risk prediction and cancer classification. Int J Mol Sci 2017; 18: 308
Address for correspondence
Publication History
Received: 27 January 2020
Accepted: 18 March 2020
Article published online:
28
June 2021
© 2020. Indian Society of Medical and Paediatric Oncology. This is an open access article published by Thieme under the terms of the Creative Commons Attribution-NonDerivative-NonCommercial-License, permitting copying and reproduction so long as the original work is given appropriate credit. Contents may not be used for commercial purposes, or adapted, remixed, transformed or built upon. (https://creativecommons.org/licenses/by-nc-nd/4.0/.)
Thieme Medical and Scientific Publishers Pvt. Ltd.
A-12, 2nd Floor, Sector 2, Noida-201301
UP, India

| Figure 1:Steps of Sanger’s sequencing
References
- Sanger F, Coulson AR. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol 1975; 94: 441-8
- Grada A, Weinbrecht K. Next-generation sequencing: Methodology and application. J Invest Dermatol 2013; 133: e11
- Heather JM, Chain B. The sequence of sequencers: The history of sequencing DNA. Genomics 2016; 107: 1-8
- Kamps R, Brandão RD, Bosch BJ, Paulussen AD, Xanthoulea S, Blok MJ. et al. Next-generation sequencing in oncology: Genetic diagnosis, risk prediction and cancer classification. Int J Mol Sci 2017; 18: 308


 PDF
PDF  Views
Views  Share
Share

