Brain and Acute Leukemia, Cytoplasmic Gene Overexpression as a Prognostic Factor in Egyptian De novo Adult Acute Myeloid Leukemia Patients
CC BY-NC-ND 4.0 · Indian J Med Paediatr Oncol 2020; 41(06): 859-868
DOI: DOI: 10.4103/ijmpo.ijmpo_215_20
Abstract
Background: Brain and acute leukemia, cytoplasmic (BAALC) gene is identified on chromosome 8q22.3 and implicated in normal hematopoiesis. BAALC gene overexpression is associated with poor outcome. Methods: We aimed to evaluate BAALC expression in de novo Egyptian acute myeloid leukemia (AML) cases and determine its prognostic value. We recruited 70 patients with de novo AML diagnosed and treated at clinical pathology and medical oncology departments, fulfilling inclusion criteria in our prospective study and evaluated BAALC expression level. Patients received induction therapy. The Institutional Review Board approved our study. Results: The mean age was 39.2 years ± 11.87, (18–60) with a male/female ratio of 3/2. The cutoff value of BAALC as a prognostic factor was 2.11 with sensitivity (86.1%), specificity (80%), positive predictive value (88.6%), and negative predictive value (76.2%.) (P < 0 class="i" xss=removed>P = 0.03). Patients with low BAALC (123.1 ± 4.9) had longer mean survival time than high BAALC group (45.85 ± 5.1) (P = 0.000). Conclusion: High-BAALC expression is an adverse prognostic factor, with a higher risk of relapse, lower CR rates, and lower survival in Egyptian de novo AML patients.
Keywords
Acute myeloid leukemia - adult - brain and acute leukemia - cytoplasmic - de novo - prognosticPublication History
Received: 05 May 2020
Accepted: 02 October 2020
Article published online:
14 May 2021
© 2020. Indian Society of Medical and Paediatric Oncology. This is an open access article published by Thieme under the terms of the Creative Commons Attribution-NonDerivative-NonCommercial-License, permitting copying and reproduction so long as the original work is given appropriate credit. Contents may not be used for commercial purposes, or adapted, remixed, transformed or built upon. (https://creativecommons.org/licenses/by-nc-nd/4.0/.)
Thieme Medical and Scientific Publishers Pvt. Ltd.
A-12, 2nd Floor, Sector 2, Noida-201301 UP, India
Abstract
Background: Brain and acute leukemia, cytoplasmic (BAALC) gene is identified on chromosome 8q22.3 and implicated in normal hematopoiesis. BAALC gene overexpression is associated with poor outcome. Methods: We aimed to evaluate BAALC expression in de novo Egyptian acute myeloid leukemia (AML) cases and determine its prognostic value. We recruited 70 patients with de novo AML diagnosed and treated at clinical pathology and medical oncology departments, fulfilling inclusion criteria in our prospective study and evaluated BAALC expression level. Patients received induction therapy. The Institutional Review Board approved our study. Results: The mean age was 39.2 years ± 11.87, (18–60) with a male/female ratio of 3/2. The cutoff value of BAALC as a prognostic factor was 2.11 with sensitivity (86.1%), specificity (80%), positive predictive value (88.6%), and negative predictive value (76.2%.) (P < 0 class="i" xss=removed>P = 0.03). Patients with low BAALC (123.1 ± 4.9) had longer mean survival time than high BAALC group (45.85 ± 5.1) (P = 0.000). Conclusion: High-BAALC expression is an adverse prognostic factor, with a higher risk of relapse, lower CR rates, and lower survival in Egyptian de novo AML patients.
Keywords
Acute myeloid leukemia - adult - brain and acute leukemia - cytoplasmic - de novo - prognosticIntroduction
Acute myeloid leukemia (AML) is a clonal myeloid neoplasm with myelopoiesis maturation arrest that leads to myeloblasts accumulation in the bone marrow (BM) and/or blood. According to the current WHO classification, myeloblasts must comprise at least 20% of nucleated cells in the BM or blood to establish a diagnosis of AML.[1]
Nowadays, therapeutic options for AML that incorporate therapies targeted at specific dysregulated genetic pathways are available. Therefore, 2008 WHO Classification of AML relies mainly on cytogenetic abnormalities and mutations in three oncogenes (NPM1, FLT3, and CEBPA) for genetic sub-classification.[1]
Deregulation expression of genes involved in cell survival, proliferation and differentiation, e.g., (brain and acute leukemia and cytoplasmic [BAALC],[2],[3] ERG, MN1 and WT1, and EVI1 were identified as the prognostic markers.[4]
BAALC gene identified on chromosome 8q22.3 is implicated in normal hematopoiesis. However, BAALC function in the hematopoietic system and its contribution in leukemogenesis are not fully understood. BAALC blocks myeloid differentiation, thus requiring a second mutation to induce leukemogenesis.[5]
BAALC overexpression is an indicator of AML aggressiveness as it is associated with poor clinical outcome.[6] High-BAALC expression is associated with lower CR rates, shorter event free survival, and shorter overall survival.[7]
Methods
Study design
We carried out this prospective cohort study in the Clinical Pathology Department, Scientific and Medical Research Center and Medical Oncology Department, Faculty of Medicine, from January 2015 and February 2019. The Faculty of Medicine Institutional Review Board and the Ethical Committee approved this study. We evaluated BAALC expression levels in acute myeloid leukemia patients and assessed its prognostic significance. A total of 70 patients were recruited, out of 107 patients in 12 months, informed consent was obtained from all patients. Furthermore, six individuals matched for age and sex were enrolled as internal control, patient's distribution is shown in [Figure 1].
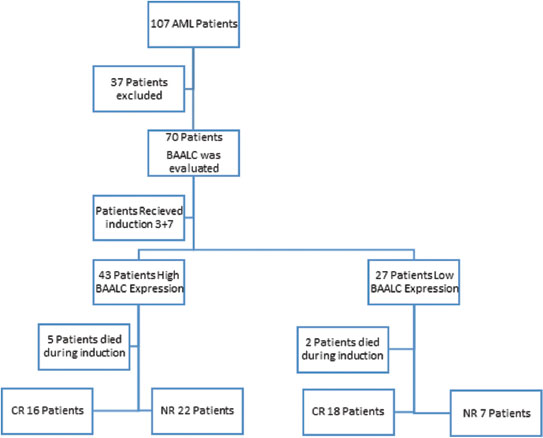
| Figure.1:Patients distribution
Patient selection
We enrolled in the study de novo adult AML patients aged >18 years old with normal liver, kidney and cardiac function tests with no concurrent malignancy and PS ≤2. We excluded patients with secondary or relapsed AML or AML (M3).
Methods of brain and acute leukemia and cytoplasmic detection
We withdrew 2 ml of venous blood aseptically from each patient by venipuncture into EDTA vacutainer for ESR. 2 ml of blood was directly used for RNA extraction.
Molecular detection of brain and acute leukemia and cytoplasmic RNA gene expression
RNA extraction from whole blood
We used Pure Link RNA Mini Kit (Life Technologies, USA) to purify RNA from anticoagulated BM sample.
Reverse-transcription polymerase chain reaction of total RNA to cDNA
To synthesize single-stranded cDNA from total RNA, we use the high capacity cDNA reverse transcription kits:
We prepared the 2 × Reverse Transcription Master Mix as per 20 μl reaction.
Real-time reverse transcription polymerase chain reaction
Using Taqman gene expression assay.
Principle
Fluorescent dye intercalates into the amplification product, which enables the rapid analysis of target DNA during the polymerase chain reaction (PCR) process.
The real-time polymerase chain reaction
Using the stratagene m × 3005p quantitative PCR system.
Interpretation of results
We normalized the transcription levels of target genes to those of B-actin (using a reference gene for accounting for the variability of cDNA amount in each sample).
Target genes expressed and presented as fold change of gene expression relative to controls. BAALC expression was positive if their expression level was one log higher than the mean level of expression reported for the control group, so the mean level for control was 1, samples were positive if expression level was more than 2.[8]
Assessment
We assessed patients demographics, clinical and laboratory data at the diagnosis (complete blood count [CBC], blood film, kidney function tests, serum uric acid, electrolytes level, liver function tests, coagulation profile, virology (HCV Ab, HBs Ag, HBc Ab, and HIV Ab)), and CSF cytological analysis (if needed). Specific investigations include BMA, immunophenotyping, karyotyping, and reverse transcriptase PCR for BAALC gene expression. We treated the patients with induction regimen (3 + 7 protocol) consisting of cytarabine (100 mg/m2/24 h) continuous infusion daily for seven consecutive days, combined with doxorubicin (30 mg/m2/24 h) for 3 days. An induction protocol consisting of (2 + 14 protocol) (cytarabine 10 mg/m2/12 h) daily for 14 days combined with 2 days of doxorubicin (25 mg/m2), was offered to patients who are over 60 years or to those who are between 55 and 60 with multiple comorbidities. We evaluated CBC and BM aspirate on day 14, 28 to evaluate the remission state.
The patient achieved CR if BM was normocellular, containing <5>15% blasts in BM), or induction death (ID: Defined as related to treatment or hypoplasia). We followed patients for 3 years to evaluate the survival.
Consolidation therapy: 3–4 cycles of high dose Ara-C (1 g/m2/12 h at days 1, 3, 5) postremission.
Statistical analysis
We used version 23.0. of IBM SPSS statistics (IBM Corp., Armonk, NY, USA); percentage analysis was done for categorical data; mean and standard deviation for continuous variables; survival was estimated using the Kaplan–Meier method. We evaluated the relationship between different prognostic and predictor variables with survival using the log-rank test. Independent prognostic variables affecting survival were evaluated using Cox-regression analysis. Tests were two-sided. P ≤ 0.05 was considered as statistically significant, P ≤ 0.001 as considered as highly significant.
Results
The present study included 70 patients. Forty-three patients (61.4%) were males, 27 females, and their ages ranged from 19 to 58 years, with a mean of 39.2 years ± 11.87.
Patients' characteristics are described in [Table 1], where 22 patients had cytogenetic abnormalities, 6 patients of M2 show t (8; 21), 7 patients of M4 show inv (16), 3 patients of M1 and 2 patient from M5 show tri (8), and 4 patients of M5 show t (11;12).
|
Variable |
AML patients (n=70), n (%) |
|---|---|
|
AML: Acute myeloid leukemia, TLC: Total leukocyte count, HB: Hemoglobin, PB: Peripheral blood, PLT: Platelet, BM: Bone marrow, ESR: Erythrocyte sedimentation rate, LDH: Lactate dehydrogenase, SD: Standard deviation, FAB: The French- American-British classification of AML |
|
|
Bone aches |
65 (92.9) |
|
Fatigue |
57 (81.4) |
|
Gum hypertrophy |
49 (70) |
|
Purpura |
51 (72.9) |
|
Bleeding |
50 (71.4) |
|
Fever |
50 (71.4) |
|
Splenomegaly |
11 (15.7) |
|
Hepatomegaly |
11 (15.7) |
|
Lymphadenopathy |
6 (8.6) |
|
TLC (x109 /L), median (range) |
48.70 (12- 161) |
|
Hb (g/dl), mean±SD (range) |
6.30±1.7 (3.4- 9.5) |
|
PLT (x109/L), median (range) |
28 (5- 100) |
|
PB blasts (%), mean±SD (range) |
58.05±17.05 (29-90) |
|
BM blasts (%), mean±SD (range) |
74.23±13.18 (52- 95) |
|
ESR (mm/h), mean±SD (range) |
111.1±17.16 (90- 150) |
|
LDH (IU/L), mean±SD (range) |
785.5±217.62 (420- 1200) |
|
FAB |
|
|
With myeloid differentiation |
21 (30) |
|
M1 |
3 (4.3) |
|
M2 |
18 (25.7) |
|
With monocytic differentiation |
49 (70) |
|
M4 |
22 (31.4) |
|
M5 |
27 (38.6) |
|
Cytogenetics |
|
|
Normal |
48 (68.6) |
|
Abnormal |
22 (31.4) |
|
Risk |
|
|
Favorable |
13 (18.6) |
|
t (8;21) |
6 (8.6) |
|
inv (16) |
7 (10) |
|
Intermediate |
53 (75.7) |
|
Normal |
48 (68.6) |
|
Tri 8 |
5 (7.1) |
|
Adverse |
|
|
t (11;12) |
4 (5.7) |
|
Variable |
Normal cytogenetic (n=48) |
Favorable (n=13) |
Intermediate and adverse (n=9) |
P |
|---|---|---|---|---|
|
AML: Acute myeloid leukemia, TLC: Total leukocyte count, HB: Hemoglobin, PB: Peripheral blood, PLT: Platelet, BM: Bone marrow, ESR: Erythrocyte sedimentation rate, LDH: Lactate dehydrogenase, SD: Standard deviation, BAALC: Brain and Acute Leukemia, Cytoplasmic. *Statistically significant |
||||
|
Hb: Mean±SD |
5.69±1.21 |
7.55±1.96 |
7.00±2.53 |
0.002* |
|
BM blast: Mean±SD |
82.13±9.86 |
61.91±9.49 |
64.17±9.66 |
0.000* |
|
LDH: Mean±SD |
873.13±209.38 |
634.55±115.63 |
586.33±33 |
0.000* |
|
ESR: Mean±SD |
118.21±18.87 |
127.55±9.53 |
127.67±11.94 |
0.172 |
|
TLC: Median (range) |
55 (15- 150) |
27 (14- 50) |
35 (12- 161) |
0.001* |
|
PLT: Median (range) |
24 (6- 92) |
72 (35- 100) |
48.50 (5- 88) |
0.000* |
|
PB blasts: Median (range) |
54 (29- 90) |
60 (29- 83) |
70 (37- 84) |
0.365 |
|
BAALC: Median (range) |
3.21 (1.05- 13.76) |
1.13 (0.80- 10.43) |
1.18 (0.86- 3.21) |
0.000* |
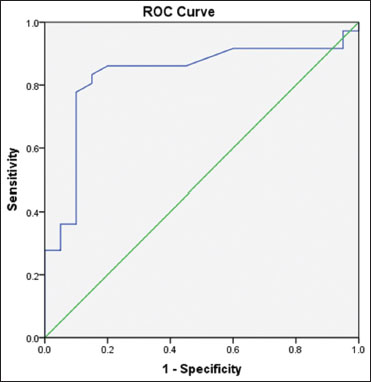
| Figure.2:Receptor operating characteristic curve for brain and acute leukemia, cytoplasmic as a prognostic factor for survival in acute myeloid leukemia patients
|
Crosstab |
||||
|---|---|---|---|---|
|
BAALC overexpression |
Variables |
Survival |
Total |
|
|
Die |
Survive |
|||
|
BAALC: Brain and acute leukemia, cytoplasmic |
||||
|
BAALC2.11 |
||||
|
>2.11 |
Count |
38 |
5 |
43 |
|
Percentage within BAALC2.11 |
84.4 |
11.6 |
100.0 |
|
|
Percentage within survival |
86.1 |
20.0 |
62.5 |
|
|
≤2.11 |
Count |
7 |
20 |
27 |
|
Percentage within BAALC2.11 |
25.9 |
74.1 |
100.0 |
|
|
Percentage within survival |
15.6 |
80.0 |
37.5 |
|
|
Total |
Count |
45 |
25 |
70 |
|
Percentage within BAALC2.11 |
64.3 |
35.7 |
100.0 |
|
|
Percentage within Survival |
100.0 |
100.0 |
100.0 |
|
|
Variable |
BAALC |
Test |
P |
|
|---|---|---|---|---|
|
>2.11 (n=43) |
≤2.11 (n=27) |
|||
|
ANOVA, t: T-test, MW: Mann- Whitney-U test. AML: Acute myeloid leukemia, TLC: Total leukocyte count, HB: Hemoglobin, PB: Peripheral blood, PLT: Platelet, BM: Bone marrow, ESR: Erythrocyte sedimentation rate, LDH: Lactate dehydrogenase, SD: Standard deviation, BAALC: Brain and Acute Leukemia, Cytoplasmic, LN: Lymphadenopathy |
||||
|
Age: Mean±SD |
38.03±12.76 |
42.24±9.96 |
t=1.29 |
0.202 |
|
Hb: Mean±SD |
5.51±1.26 |
7.33±1.77 |
t=4.46 |
0.000* |
|
BM blasts: Mean±SD |
85.20±6.09 |
61.29±6.27 |
t=14.06 |
0.000* |
|
LDH: Mean±SD |
924.74±164.47 |
580.19±79.66 |
t=8.96 |
0.000* |
|
ESR: Mean±SD |
121.72±17.26 |
119.85±35 |
t=0.38 |
0.699 |
|
TLC: Median (range) |
60 (33- 161) |
29 (12- 52) |
MW=4.68 |
0.000* |
|
Plt: Median (range) |
20 (5- 40) |
60 (30- 100) |
MW=5.90 |
0.000* |
|
PB blasts: Median (range) |
58 (29- 90) |
60 (29- 84) |
MW=0.24 |
0.806 |
|
Sex, n (%) |
||||
|
Female (27) |
17 (63) |
10 (37) |
X2=0.02 |
0.888 |
|
Male (43) |
26 (60.5) |
17 (39.5) |
||
|
Fatigue, n (%) |
||||
|
Yes(57) |
35 (61.4) |
22 (38.6) |
Fisher exact test |
1 |
|
No (13) |
8 (61.5) |
5 (38.5) |
||
|
Fever, n (%) |
||||
|
Yes(50) |
31 (62) |
19 (38) |
X2=0.141 |
0.708 |
|
No (20) |
12 (60) |
8 (40) |
||
|
Bone ache, n (%) |
||||
|
Yes(64) |
40 (64.5) |
24 (37.5) |
X2=0.287 |
0.592 |
|
No (6) |
3 (50) |
3 (50) |
||
|
Gum hypertrophy, n (%) |
||||
|
Yes (49) |
41 (83.5) |
8 (16.5) |
X2=27.25 |
0.000* |
|
No (21) |
2 (6.7) |
19 (93.3) |
||
|
Bleeding, n (%) |
||||
|
Yes (50) |
36 (72) |
14 (28) |
X2=5.97 |
0.015* |
|
No (20) |
7 (37.5) |
13 (62.5) |
||
|
Purpura, n (%) |
||||
|
Yes (51) |
36 (70.6) |
15 (29.4) |
X2=7.4 |
0.006* |
|
No (19) |
7(36.8) |
12 (63.2) |
||
|
Splenomegaly, n (%) |
||||
|
Yes(11) |
7 (63.6) |
4 (36.4) |
X2=0.07 |
0.778 |
|
No (59) |
36(61) |
23 (39) |
||
|
Hepatomegaly, n (%) |
||||
|
Yes(11) |
7 (63.6) |
4 (36.4) |
X2=0.07 |
0.778 |
|
No (59) |
36(61) |
23 (39) |
||
|
LN, n (%) |
||||
|
Yes (6) |
4 (66.7) |
2 (33.3) |
X2=0.71 |
0.397 |
|
No (64) |
39 (60.9) |
25 (39.1) |
||
|
Cytogenetics, n (%) |
||||
|
Abnormal (22) |
3 (13.6) |
19 (86.4) |
X2=26.80 |
0.000* |
|
Normal (48) |
40 (83.3) |
8 (16.7) |
||
|
Induction protocol (70), n (%) |
0.33 |
|||
|
3+7 (66 [94.3%]) |
39 (90.7) |
27 (100) |
||
|
2+14 (4[5.7%]) |
4 (9.3) |
0 (0) |
||
|
Total 70 (100%) |
43 (100) |
27(100) |
||
|
BMD14 |
0.22 |
|||
|
Evaluated patients (65[92.9%]) |
||||
|
Unevaluated patients (5[7.1%]) |
||||
|
Blasts % (median, range) (0- 40) |
1.5 (0- 12) |
5 (0- 40) |
||
|
BMD28 |
||||
|
Evaluated patients [63 (90%)] |
||||
|
Unevaluated (7 [10%]) |
||||
|
Cellularity, n (%) |
0.88 |
|||
|
Normocellular (39[62%]) |
24 (63.1) |
15 (60) |
||
|
Hypocellular (12 [19%]) |
7 (18.4) |
5 (20) |
||
|
Hypercellular (12 [19%]) |
7 (18.4) |
5 (20) |
||
|
Total |
38 (100) |
25 (100) |
0.07 |
|
|
Blasts % (median, range) 2 (0- 21) Induction response (n=63), n (%) |
2 (0-21) |
3 (1-20) |
||
|
CR (34[54%]) |
16 (42.1) |
18 (72) |
0.03 |
|
|
Not in CR (29 [46%]) |
22 (57.9) |
7 (28) |
||
|
Total |
38 (100) |
25 (100) |
||
AUC=0.83. CI=0.71- 0.94. CI: Confidence interval, SE: Standard error, BAALC: Brain And Acute Leukemia, Cytoplasmic, AUC: Area under the curve
|
AUC |
||||
|---|---|---|---|---|
|
Test result variable(s): BAALC |
||||
|
Area |
SE |
Asymptotic significance |
Asymptotic 95% CI |
|
|
Lower bound |
Upper bound |
|||
|
0.832 |
0.060 |
0.000 |
0.715 |
0.949 |
Factors that affected survival significantly on univariate analysis were entered for multivariate analysis in cox-proportional hazard model. There was a statistically significant association between survival and gum hypertrophy, i.e., patients with gum hypertrophy are at a risk of dying 2.2 more than those with no gum hypertrophy, cytogenetics, i.e., patients with abnormal cytogenetics carry less risk 0.45 to die than those with normal cytogenetics, laboratory data, i.e., patients with low hemoglobin level, increased BM blasts, increased LDH level, higher TLC, lower PLT count, increased PB blasts, and high BAALC expression are at risk to die [Table 5].
|
Variable |
Survival |
P±RR (95% CI) |
|
|---|---|---|---|
|
Die (45) |
Survive (25) |
||
|
t: T-test, MW: Mann- Whitney-U test. AML: Acute myeloid leukemia, TLC: Total leukocyte count, HB: Hemoglobin, PB: Peripheral blood, PLT: Platelet, BM: Bone marrow, ESR: Erythrocyte sedimentation rate, LDH: Lactate dehydrogenase, SD: Standard deviation, BAALC: Brain and Acute Leukemia, Cytoplasmic, CI: Confidence interval, RR: Relative risk, LN: Lymphadenopathy. *Statistically significan |
|||
|
Sex, n (%) |
|||
|
Female (27) |
19 (70.4) |
8 (29.6) |
0.625 RR 1.1 (0.74- 1.62) |
|
Male (43) |
26 (60.5) |
17 (39.5) |
|
|
Fatigue, n (%) |
|||
|
Yes(57) |
36 (63.2) |
21 (36.8) |
0.96 RR 1.0 (0.61- 1.66) |
|
No (13) |
9 (69) |
4 (31) |
|
|
Fever, n (%) |
|||
|
Yes (50) |
32 (64) |
18 (36) |
0.96 RR 0.99 (0.64- 1.51) |
|
No (20) |
13(65) |
7 (35) |
|
|
Bone ache, n (%) |
|||
|
Yes (64) |
42 (65.6) |
22 (34.4) |
0.53 RR 1.3 (0.48- 3.55) |
|
No (6) |
3 (50) |
3 (50) |
|
|
Gum hypertrophy, n (%) |
|||
|
Yes (49) |
37 (75.5) |
12 (24.5) |
0.003* RR 2.2 (1.08- 4.73) |
|
No (21) |
8 (38) |
13(62) |
|
|
Bleeding, n (%) |
|||
|
Yes (50) |
33 (66) |
17(34) |
0.86 RR 1.04 (0.66- 1.61) |
|
No (20) |
12 (60) |
8 (40) |
|
|
Purpura, n (%) |
|||
|
Yes (51) |
33 (64.8) |
18 (35.2) |
0.68 RR 1.09 (0.68- 1.75) |
|
No (19) |
12 (63.2) |
7 (36.8) |
|
|
Splenomegaly, n (%) |
|||
|
Yes(11) |
7 (63.6) |
4 (36.4) |
0.87 RR 1.04 (0.62- 1.73) |
|
No (59) |
38 (64.4) |
21 (35.6) |
|
|
Hepatomegaly, n (%) |
|||
|
Yes(11) |
7(63.6) |
4 (36.4) |
0.87 RR 1.04 (0.62- 1.73) |
|
No (59) |
38 (64.4) |
21 (35.6) |
|
|
LN, n (%) |
|||
|
Yes (6) |
4 (66.7) |
2 (33.3) |
0.44 RR 1.27 (0.78- 2.07) |
|
No (64) |
41 (64.1) |
23 (35.9) |
|
|
Cytogenetics, n (%) |
|||
|
Abnormal (22) |
8 (36.4) |
14 (63.6) |
0.000* RR 0.45 (0.23- 0.89) |
|
Normal (48) |
37 (77.1) |
11(22.9) |
|
|
Hb (g/dl): Mean±SD |
5.72±1.4 |
7.05±1.82 |
0.004* |
|
BM blasts: Mean±SD |
81.6±11.01 |
66.10±10.5 |
0.000* |
|
ESR: Mean±SD |
121.72±17.26 |
119.85±35 |
0.699 |
|
LDH: Mean±SD |
868.75±204.24 |
663.75±178.35 |
0.000* |
|
TLC: (x109/L), median (range) |
56 (14- 161) |
42.50 (12- 136) |
0.006* |
|
PLT: (x109/L), median (range) |
24 (5- 95) |
44 (12- 100) |
0.001* |
|
PB blasts: Median (range) |
58.50 (29- 90) |
59 (29- 84) |
0.000* |
|
BAALC: Median (range) |
3.32 (0.80¬13.7) |
1.21 (0.86- 6.88) |
0.000* |
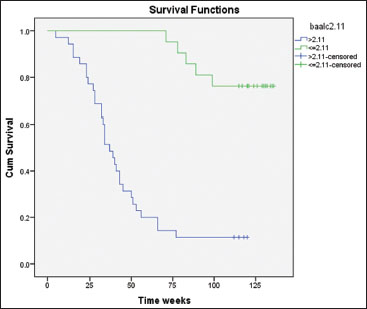
| Figure.3:Cumulative survival time according to brain and acute leukemia, cytoplasmic among studied acute myeloid leukemia patients by Kaplan– Meier test
|
Total (n) |
Death |
Censored (still alive),n(%) |
Mean survival time (weeks) X±SD |
95% CI |
Log-rank test |
P |
|
|---|---|---|---|---|---|---|---|
|
CI: Confidence interval, BAALC: Brain and Acute Leukemia, Cytoplasmic, AML: Acute myeloid leukemia, SD: Standard deviation |
|||||||
|
Chemotherapy response |
|||||||
|
No CR |
29 |
29 |
0 (0.0) |
34.2±2.8 |
28.74- 39.73 |
62.62 |
0.000 |
|
CR |
34 |
7 |
27 (79.4) |
121.1±4.9 |
111.48- 130.88 |
||
|
Overall |
63 |
36 |
27 (42.9) |
||||
|
Cytogenetics |
|||||||
|
CN-AML |
43 |
30 |
13 (30.2) |
60.25±7.13 |
46.28- 74.23 |
10.83 |
0.004 |
|
Favorable |
13 |
2 |
11 (84.6) |
114.00±8.64 |
97.05- 130.94 |
||
|
Inter and poor |
7 |
4 |
3 (42.9) |
107.16±16.71 |
74.40- 139.92 |
||
|
Overall |
63 |
36 |
27 (42.9) |
||||
|
BAALC2.11 |
|||||||
|
>2.11 |
39 |
32 |
7 (17.9) |
45.85±5.1 |
35.66- 56.04 |
31.20 |
0.000* |
|
≤2.11 |
34 |
4 |
20 (83.3) |
123.61±4.9 |
113.93- 133.29 |
||
|
Overall |
63 |
36 |
27 (42.9) |
||||
|
Toxicity type |
During induction (n=63), n (%) |
During consolidation (n=56), n (%) |
|---|---|---|
|
According to CTCAE Version 5.0. CTCAE: Common Terminology Criteria for Adverse Events |
||
|
Hematological toxicity Neutropenia |
||
|
G3 |
0 (0) |
4 (7) |
|
G4 |
63(100) |
52 (93) |
|
Anemia |
||
|
G3 |
53 (84.1) |
45 (80) |
|
G4 |
10 (15.9) |
11 (20) |
|
Thrombocytopenia |
||
|
G3 |
0 (0) |
3 (5.4) |
|
G4 |
63 (100) |
53 (94.6) |
|
Fever and infection Neutropenic fever |
||
|
G3 |
55 (87.3) |
45 (80) |
|
G4 |
8 (12.7) |
11 (20) |
|
Cannula site infection |
||
|
G2 |
0 (0) |
4 (7) |
|
G3 |
3 (4.8) |
3 (5.4) |
|
G4 |
1 (16) |
1 (18) |
|
Sepsis |
||
|
G3 |
1 (16) |
4 (7) |
|
G4 |
7 (11.1) |
1 (18) |
|
Chest infection |
||
|
G3 |
9 (14.3) |
12 (21.4) |
|
G4 |
3 (4.8) |
0 (0) |
|
Soft-tissue infection |
||
|
G3 |
1 (16) |
2 (3.6) |
|
G4 |
2 (3.2) |
1 (18) |
|
Typhlitis |
||
|
G3 |
4 (6.3) |
2 (3.6) |
|
G4 |
2 (3.2) |
1 (18) |
|
Fungal infection |
||
|
G3 |
2 (3.2) |
7 (12.5) |
|
G4 |
6 (9.5) |
4 (7) |
|
Other toxicities Gastritis |
||
|
G2 |
15 (23.8) |
8 (14.3) |
|
G3 |
12 (19) |
12 (21.4) |
|
Vomiting |
||
|
G2 |
4 (6.3) |
4 (7) |
|
G3 |
2 (3.2) |
2 (3.6) |
|
Diarrhea |
||
|
G2 |
3 (4.8) |
4 (7) |
|
G3 |
2 (3.2) |
0 (0) |
|
Liver enzymes increased |
||
|
G2 |
2 (3.2) |
5 (8.9) |
|
G3 |
2 (3.2) |
1 (18) |
|
Blood bilirubin |
||
|
increased |
||
|
G2 |
2 (3.2) |
2 (3.6) |
|
G3 |
3 (4.8) |
0 (0) |

| Figure.1:Patients distribution

| Figure.2:Receptor operating characteristic curve for brain and acute leukemia, cytoplasmic as a prognostic factor for survival in acute myeloid leukemia patients

| Figure.3:Cumulative survival time according to brain and acute leukemia, cytoplasmic among studied acute myeloid leukemia patients by Kaplan– Meier test
References
- Vardiman JW, Arber DA, Brunning RD, Larson RA, Matutes E, Baumann I. et al. Therapy-related myeloid neoplasms. In: Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H. et al.editors WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon: IARC; 2008. p. 127-9
- Baldus CD, Thiede C, Soucek S, Bloomfield CD, Thiel E, Ehninger G. BAALC expression and FLT3 internal tandem duplication mutations in acute myeloid leukemia patients with normal cytogenetics: Prognostic implications. J Clin Oncol 2006; 24: 790-7
- Langer C, Radmacher MD, Ruppert AS, Whitman SP, Paschka P, Mrózek K. et al. High BAALC expression associates with other molecular prognostic markers, poor outcome, and a distinct gene-expression signature in cytogenetically normal patients younger than 60 years with acute myeloid leukemia: A cancer and leukemia Group B (CALGB) study. Blood 2008; 111: 5371-9
- Metzeler KH, Dufour A, Benthaus T, Hummel M, Sauerland MC, Heinecke A. et al. ERG expression is an independent prognostic factor and allows refined risk stratification in cytogenetically normal acute myeloid leukemia: A comprehensive analysis of ERG, MN1, and BAALC transcript levels using oligonucleotide microarrays. J Clin Oncol 2009; 27: 5031-8
- Heuser M, Berg T, Kuchenbauer F, Lai CK, Park G, Fung S. et al. Functional role of BAALC in leukemogenesis. Leukemia 2012; 26: 532-6
- Franzoni A, Passon N, Fabbro D, Tiribelli M, Damiani M, Damante G, Sarin YK. Histone post-translational modifications associated to BAALC expression in leukemic cells. Biochem Biophys Res Commun 2012; 417: 721-5
- Weber S, Alpermann T, Dicker F, Jeromin S, Nadarajah N, Eder C. et al. BAALC expression: A suitable marker for prognostic risk stratification and detection of residual disease in cytogenetically normal acute myeloid leukemia. Blood Cancer J 2014; 10: e173
- Spanaki A, Perdikogianni C, Linardakis E, Kalmanti M. Quantitative assessment of PRAME expression in diagnosis of childhood acute leukemia. Leuk Res 2007; 31: 639-42
- Baldus CD, Tanner SM, Ruppert AS, Whitman SP, Archer KJ, Marcucci G. et al. BAALC expression predicts clinical outcome of de novo acute myeloid leukemia patients with normal cytogenetics: A cancer and leukemia Group B study. Blood 2003; 102: 1613-8
- Bienz M, Ludwig M, Leibundgut EO, Mueller BU, Ratschiller D, Solenthaler M. et al. Risk assessment in patients with acute myeloid leukemia and a normal karyotype. Clin Cancer Res 2005; 11: 1416-24
- Rashed R, Kadry D, EL Taweel M, El Wahab NA, El TA.Hameed. Relation of BAALC and ERG gene expression with overall survival in acute myeloid leukemia cases. Asian Pac J Cancer Prev 2015; 16: 7875-82
- Amirpour M, Ayatollahi H, Sheikhi M, Azarkerdar S, Shams SF. Evaluation of BAALC gene expression in normal cytogenetic acute myeloid leukemia patients in North-East of Iran. Med J Islam Repub Iran 2016; 30: 418
- Zhou JD, Yang L, Zhang YY, Yang J, Wen XM, Guo H. et al. Overexpression of BAALC: Clinical significance in Chinese de novo acute myeloid leukemia. Med Oncol 2015; 32: 386
- Soliman A, Aal AA, Afify R, Ibrahim N. BAALC and ERG expression in Egyptian patients with acute myeloid leukemia, relation to survival and response to treatment. Open Access Maced J Med Sci 2016; 4: 264-70
- Qi X, Shen Y, Cen J, Chen H, Sun Y, Sheng H. et al. Up-regulation of BAALC gene may be an important alteration in AML-M2 patients with t (8; 21) translocation. J Cell Mol Med 2008; 12: 2301-4
- Schwind S, Marcucci G, Maharry K, Radmacher MD, Mrózek K, Holland KB. et al. BAALC and ERG expression levels are associated with outcome and distinct gene and microRNA expression profiles in older patients with de novo cytogenetically normal acute myeloid leukemia: A cancer and leukemia Group B study. Blood 2010; 116: 5660-9
- El-Sharnouby JA, Ahmed LM, Taha AM, Kamal O. Prognostic significance of CEBPA mutations and BAALC expression in acute myeloid leukemia Egyptian patients with normal karyotype. Egypt J Immunol 2008; 15: 131-43
- Yahya RS, Sofan MA, Abdelmasseih HM, Saudy N, Sharaf-Eldein MA. et al. Prognostic implication of BAALC gene expression in adult acute myeloid leukemia. Clin Lab 2013; 59: 621-8
- Langer C, Marcucci G, Holland KB, Radmacher MD, Maharry K, Paschka P. et al. Prognostic importance of MN1 transcript levels, and biologic insights from MN1-associated gene and microRNA expression signatures in cytogenetically normal acute myeloid leukemia: A cancer and leukemia Group B study. J Clin Oncol 2009; 27: 3198-204
- Eisfeld AK, Marcucci G, Liyanarachchi S, Döhner K, Schwind S, Maharry K. et al. Heritable polymorphism predisposes to high BAALC expression in acute myeloid leukemia. Proc Natl Acad Sci USA 2012; 109: 6668-73


 PDF
PDF  Views
Views  Share
Share

